9.1. General considerations
A growing number of pharmacoepidemiological studies use data from networks of databases, often from different countries. Pooling data across different databases affords insight into the generalisability of the results and may improve precision. Some of these networks are based on long-term contracts with selected partners and are very well structured (such as Sentinel, the Vaccine Safety Datalink (VSD), the Canadian Network for Observational Drug Effect Studies (CNODES)) and the recently set-up Data Analysis and Real World Interrogation Network (DARWIN EU®). Others are collaborations based on open science principles such as the Observational Health Data Sciences and Informatics (OHDSI) program.
In Europe, collaborations for multi-database studies have been strongly encouraged as part of the drug safety research funded by the European Commission (EC) as well as public-private partnerships such as the Innovative Medicines Initiative (IMI). This funding resulted in the conduct of groundwork necessary to overcome the hurdles of data sharing across countries for specific projects (e.g. PROTECT, ADVANCE, EMIF, EHDEN, ConcePTION) and specific post-authorisation studies. The European Commission is currently establishing an European Health Data Space (EHDS) and major breakthroughs in this field are expected, with the Joint Action Towards the European Health Data Space – TEHDAS developing joint European principles for the secondary use of health data.
The 2009 H1N1 influenza pandemic (see Safety monitoring of Influenza A/H1N1 pandemic vaccines in EudraVigilance, Vaccine 2011;29(26):4378-87) and the 2020 COVID-19 pandemic showed the value of an operational infrastructure to rapidly and effectively monitor the safety of therapeutics and vaccines. In this context, EMA established contracts with academic and private partners to support readiness of research networks to perform observational research. Three dedicated projects started in 2020: ACCESS (vACcine Covid-19 monitoring readinESS), CONSIGN (COVID-19 infectiOn aNd medicineS In preGNancy) and E-CORE (Evidence for COVID-19 Observational Research Europe). Other initiatives have emerged to address specific COVID-19 related research questions, such as the CVD-COVID-UK consortium (see Linked electronic health records for research on a nationwide cohort of more than 54 million people in England: data resource, BMJ. 2021;373:n826), providing a secure access to linked health data from primary and secondary care, registered deaths, COVID-19 laboratory data, vaccination data and cardiovascular specialist audits. Similarly, linked data have been made available in trusted research environments in Scotland and Wales.
EMA funded several studies to address research questions on the monitoring of COVID-19 vaccines using federated analytics with a common data model (CDM), which resulted in publications on background rates of adverse events of special interest (Characterising the background incidence rates of adverse events of special interest for covid-19 vaccines in eight countries: multinational network cohort study, BMJ. 2021;373:n1435; Background rates of 41 adverse events of special interest for COVID-19 vaccines in 10 European healthcare databases - an ACCESS cohort study, Vaccine. 2023;41(1):251-262); thrombosis and risk of coagulopathy post-COVID-19 (Venous or arterial thrombosis and deaths among COVID-19 cases: a European network cohort study, Lancet Infectious Diseases 2022;22(8):1142-52); comparative risk of thrombosis and thrombocytopenia following COVID-19 vaccines (Comparative risk of thrombosis with thrombocytopenia syndrome or thromboembolic events associated with different covid-19 vaccines: international network cohort study from five European countries and the US, BMJ. 2022;379:e071594); and myocarditis (Myocarditis and pericarditis associated with SARS-CoV-2 vaccines: A population-based descriptive cohort and a nested self-controlled risk interval study using electronic health care data from four European countries, Front Pharmacol. 2022;13:1038043).
In this Chapter, the term networking is used to reflect collaboration between researchers for sharing expertise and resources. The ENCePP Database of Research Resources, which provides an inventory of research centres and data sources collaborating on specific pharmacoepidemiology and pharmacovigilance studies in Europe, may facilitate such networking by allowing the identification of research centres and data sources by country, study, type of research, and other relevant fields.
The use of research networks in medicines safety and utilisation, and in disease epidemiology, is well established, with a significant body of practical experience. Their use in effectiveness research is now increasing (see Assessing strength of evidence for regulatory decision making in licensing: What proof do we need for observational studies of effectiveness?, Pharmacoepidemiol Drug Saf. 2020;29(10):1336-40).
From a methodological point of view, studies adopting a multi-database design have many advantages over single database studies:
-
It increases the size of the study population. This especially facilitates research on rare events, on medicines used in specialised settings (see Ability of primary care health databases to assess medicinal products discussed by the European Union Pharmacovigilance Risk Assessment Committee, Clin Pharmacol Ther. 2020;107(4):957-65), or when the interest is in subgroup effects.
-
It exploits the heterogeneity of treatment options across countries, which allows studying the effect of different medicines used for the same indication, or specific patterns of utilisation.
-
It exploits differences in outcome/event rates across countries/regions.
-
It provides additional knowledge on the generalisability of results and on the consistency of associations, for instance whether a safety issue can be identified in several countries. Possible inconsistencies might be caused by different biases or truly different effects in the databases, revealing causes of differential effects, and these might be investigated.
-
It involves experts from various countries addressing case definitions, terminologies, coding in databases, and research practices. This provides opportunities to increase consistency of results of observational studies.
-
For primary data collection from multiple data sources, it shortens the time needed for obtaining the desired sample size and therefore accelerates the investigation of safety issues or other outcomes.
The articles Approaches for combining primary care electronic health record data from multiple sources: a systematic review of observational studies (BMJ Open 2020;10(10): e037405) and Different strategies to execute multi-database studies for medicines surveillance in real world setting: a reflection on the European model (Clin Pharmacol Ther. 2020;108(2):228-35) describe key characteristics of studies using multiple data sources and different models applied for combining data or results from multiple databases. A common characteristic of all models is the fact that data partners maintain physical and operational control over electronic data in their existing environment, and therefore, the data extraction is always performed locally. Differences, however, exist in the following areas: use of a common protocol; use of a CDM; and where and how the data analysis is conducted.
Use of a CDM implies that local formats are translated into a predefined, common data structure, which allows launching a similar data extraction and analysis script across several databases. Sometimes the CDM also imposes a common terminology, such as for the OMOP CDM. The CDM can be systematically applied on the entire database (generalised CDM) or on the subset of data needed for a specific study (study-specific CDM). While transforming the database in a CDM, comparisons between source and target data across all variables and dimensions is strongly recommended as part of the quality control of the process, in order to make sure that the transformation faithfully represents the source data, both in terms of completeness and accuracy. A number of tools exist for checking the resulting data, including the OHDSI DataQualityDashboard, which involves thousands of checks for conformance, completeness, and plausibility, based on the harmonised framework for data quality assessment developed by Khan et al. (EGEMS 2016;4(1):1244).
In the European Union, study specific CDMs have generated results for several projects, and several databases have been converted to a generalised CDM version that exists alongside the native version. This conversion was accelerated as a result of the observational research needed to respond to the COVID-19 pandemic. An example of application of generalised CDMs are studies conducted in the OHDSI community, such as Association of angiotensin converting enzyme (ACE) inhibitors and angiotensin 2 receptor blockers (ARB) on COVID-19 incidence and complications or the ConcePTION study From Inception to ConcePTION: Genesis of a Network to Support Better Monitoring and Communication of Medication Safety During Pregnancy and Breastfeeding (Clin Pharmacol Ther. 2022;111(1):321-31). More recently, DARWIN EU® has galvanised the use of the OMOP CDM for regulatory purposes, with the completion of the first studies, and the planned commissioning of many additional studies in the coming years (see list of completed DARWIN EU® studies).
9.2. Models of studies using multiple data sources
Studies may be classified into five categories according to specific choices in the steps needed for their execution, i.e., protocol and statistical analysis plan (SAP) development, location of data extraction and analysis (locally or centrally), methods for data extraction and analysis (using individual or common programs, use of a CDM, and which type of CDM: study-specific or general CDM). The key steps needed to execute each study model are presented in the following Figure and explained in this section.
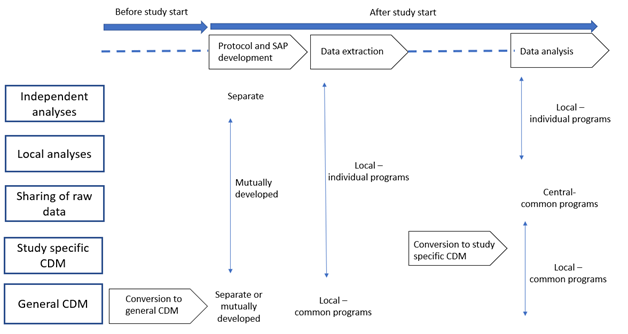
9.2.1. Independent analyses: separate protocols, local and individual data extraction and analysis, no CDM
The traditional model to combine data from multiple data sources consists in data extraction and analysis performed independently at each centre, based on separate protocols. This is usually followed by a meta-analysis of the different estimates obtained (see Chapter 10 and Annex 1).
This type of model, when viewed as a means to combine results from multiple data sources on the same research questions, may be considered as a baseline situation which a research network should try to improve on for the study design. Meta-analyses also facilitate the evaluation of heterogeneity of results across different independent studies and they could be performed retrospectively regardless of the model of studies used, in line with the recommendations from the Multi-centre, multi-database studies with common protocols: lessons learnt from the IMI PROTECT project (Pharmacoepidemiol Drug Saf. 2016;25(S1):156-65). Investigating heterogeneity may provide useful information on the issue under investigation, and explaining such variation should also be attempted if the data sources can be accessed. An example of such an investigation is Assessing heterogeneity of electronic health-care databases: A case study of background incidence rates of venous thromboembolism (Pharmacoepidemiol Drug Saf. 2023 Apr 17. doi: 10.1002/pds.5631). This approach increases consistency in findings from observational drug effect studies or reveals causes of differential drug effects.
9.2.2. Local analysis: common protocol, local and individual data extraction and analysis, no CDM
In this model, data are extracted and analysed locally, with site-specific programs developed by each centre, on the basis of a common protocol and a common SAP agreed by all study partners. The common SAP defines and standardises exposures, outcomes and covariates, analytical programmes and reporting formats. The results of each analysis, either at the subject level or in an aggregated format depending on the governance of the network, are shared and can be pooled together using meta-analysis.
This approach allows the assessment of database or population characteristics and their impact on estimates, but it reduces the variability of results determined by differences in design. Examples of research networks that use the common protocol approach are PROTECT (as described in Improving Consistency and Understanding of Discrepancies of Findings from Pharmacoepidemiological Studies: the IMI PROTECT Project, Pharmacoepidemiol Drug Saf. 2016;25(S1): 1-165), which has implemented this approach in collaboration with CNODES (see Major bleeding in users of direct oral anticoagulants in atrial fibrillation: A pooled analysis of results from multiple population-based cohort studies, Pharmacoepidemiol Drug Saf. 2021;30(10):1339-52).
This approach requires very detailed common protocols and data specifications that reduce variability in interpretation by researchers.
9.2.3. Sharing of data: common protocol, local and individual data extraction, central analysis
In this approach, a common protocol is agreed by the study partners. Data intended to be used for the study are locally extracted with site-specific programs, transferred without analysis and conversion to a CDM, and pooled and analysed at the central partner receiving them. Data received at the central partner can be reformatted to a common structure to facilitate the analysis.
This approach applies when databases are very similar in structure and content, as for some Nordic registries and the Italian regional databases. Examples of such models are Protocol: Methodology of the brodalumab assessment of hazards: a multicentre observational safety (BRAHMS) study (BMJ. Open 2023;13(2):e066057) and All‐cause mortality and antipsychotic use among elderly persons with high baseline cardiovascular and cerebrovascular risk: a multi‐center retrospective cohort study in Italy (Expert Opin. Drug Metab. Toxicol. 2019;15(2):179-88).
The central analysis allows for assessment of pooled data adjusting for covariates on an individual patient level and removing an additional source of variability linked to the statistical programming and analysis. However, this model becomes more difficult to implement due to the stronger privacy requirements for sharing patient level data.
9.2.4. Study specific CDM: common protocol, local and individual data extraction, local and common analysis, study specific CDM
In this approach, a common protocol is agreed by the study partners. Data intended to be used for the study are locally extracted and transformed into an agreed CDM. The data in the CDM are then processed locally in every site with one common program. The output of the common program is transferred to a specific partner. The output to be shared may be an analytical dataset or study estimates, depending on the governance of the network. This model is explained in From Inception to ConcePTION: Genesis of a Network to Support Better Monitoring and Communication of Medication Safety During Pregnancy and Breastfeeding (Clin Pharmacol Ther. 2022;111(1):321-31).
Examples of research networks that used this approach by employing a study-specific CDM with transmission of anonymised patient-level data (allowing a detailed characterisation of each database) are EU-ADR (as explained in Combining multiple healthcare databases for postmarketing drug and vaccine safety surveillance: why and how?, J Intern Med 2014;275(6):5511) SOS ARITMO, SAFEGUARD, GRIP, EMIF, EUROmediCAT, ADVANCE, VAC4EU and ConcePTION. In all these projects, a CDM was utilised, and R, SAS, STATA or Jerboa scripts used to create and share common analytics. Diagnosis codes for case finding can be mapped across terminologies by using the Codemapper developed in ADVANCE (see CodeMapper: semiautomatic coding of case definitions, Pharmacoepidemiol Drug Saf. 2017;26(8):998-1005). An example of a study performed using this model is Background rates of 41 adverse events of special interest for COVID-19 vaccines in 10 European healthcare databases - an ACCESS cohort study, Vaccine. 2023;41(1):251-262).
9.2.5. General CDM: common protocol, local and common data extraction and analysis, general CDM
In this approach, the local databases are transformed into a CDM prior to, and are agnostic to, any study protocol. When a study is required, a common protocol is developed and a centrally created analysis program is created that runs locally on each database to extract and analyse the data. The output of the common programs shared may be an analytical dataset or study estimates, depending on the governance of the network.
Examples of research networks which use a generalised CDM are the Sentinel Initiative (as described in The US Food and Drug Administration Sentinel System: a national resource for a learning health system, Journal of the American Medical Informatics Association, Volume 29, December 2022, Pages 2191–2200) OHDSI – Observational Health Data Sciences and Informatics, the Canadian Network for Observational Drug Effect Studies (CNODES), and EMA’s Data Analysis and Real World Interrogation Network (DARWIN EU®). The latter uses the same CDM as OHDSI, and combines previously existing analytical pipelines with bespoke newly developed ones, based on an EMA-endorsed catalogue of Standardised Analytics.
The main advantage of a general CDM is that it can be used for nearly any study involving the same database converted into the CDM. OHDSI and DARWIN EU® are based on the Observational Medical Outcomes Partnership (OMOP) CDM which is now used by many organisations and has been tested for its suitability for safety studies (see, for example, Validation of a common data model for active safety surveillance research, J Am Med Inform Assoc. 2012;19(1):54–60; and Can We Rely on Results From IQVIA Medical Research Data UK Converted to the Observational Medical Outcome Partnership Common Data Model?: A Validation Study Based on Prescribing Codeine in Children, Clin Pharmacol Ther. 2020;107(4):915-25). Conversion into the OMOP CDM requires formal mapping of database items to standardised concepts, which is a resource intensive and iterative process. Iterations on the same databases usually lead to gains in efficiency. Mapping expertise and software are also constantly developed to support and accelerate the conversion process. Examples of studies performed with the OMOP CDM in Europe are Large-scale evidence generation and evaluation across a network of databases (LEGEND): assessing validity using hypertension as a case study (J Am Med Inform Assoc. 2020;27(8):1268-77); Safety of hydroxychloroquine, alone and in combination with azithromycin, in light of rapid wide-spread use for COVID-19: a multinational, network cohort and self-controlled case series study (Lancet Rheumatol. 2020;11(2):e698–711); Characterising the background incidence rates of adverse events of special interest for covid-19 vaccines in eight countries: multinational network cohort study (BMJ. 2021;373:n1435); Venous or arterial thrombosis and deaths among COVID-19 cases: a European network cohort study (Lancet Infectious Diseases 2022;22(8):P1142-52); and Comparative risk of thrombosis with thrombocytopenia syndrome or thromboembolic events associated with different covid-19 vaccines: international network cohort study from five European countries and the US (BMJ. 2022;379:e071594).
In A Comparative Assessment of Observational Medical Outcomes Partnership and Mini-Sentinel Common Data Models and Analytics: Implications for Active Drug Safety Surveillance (Drug Saf. 2015;38(8):749-65), it is suggested that slight conceptual differences between the Sentinel and the OMOP models do not significantly impact on identifying known safety associations. Differences in risk estimations can be primarily attributed to the choices and implementation of the analytic approach.
A review of IT-architecture, legal considerations, and statistical methods for federated analyses is presented in Federated analyses of multiple data sources in drug safety studies (Pharmacoepidemiol Drug Saf. 2023 Mar;32(3):279-286).
9.3 Challenges of different models
The different models described above present several challenges, as detailed below.
Related to the database content:
-
Differences in the underlying health care systems,
-
Different mechanisms of data generation and collection as well as data availability,
-
Mapping of different drugs and disease dictionaries (e.g., SNOMED, the International Classification of Disease, 10th Revision (ICD-10), Read codes),
-
Free text medical notes in different languages,
-
Differences in the validation of study variables and access to source documents for validation,
-
Differences in the type and quality of information contained within each database.
Related to the organisation of the network:
-
Different ethical and governance requirements in each country regarding processing of anonymised or pseudo-anonymised healthcare data,
-
Issues linked to intellectual property and authorship,
-
Implementing quality controls procedures at each partner and across the entire network,
-
Sustainability and funding mechanisms,
-
The networks tend to become very topic specific over time and to become isolated in ‘silos’.
Each model has strengths and weaknesses in facing the above challenges, as illustrated in Data Extraction and Management in Networks of Observational Health Care Databases for Scientific Research: A Comparison of EU-ADR, OMOP, Mini-Sentinel and MATRICE Strategies (eGEMs 2016;4(1):2). In particular, a central analysis or a CDM provide protection from problems related to variation in how protocols are implemented by individual analysts (as described in Quantifying how small variations in design elements affect risk in an incident cohort study in claims; Pharmacoepidemiol Drug Saf. 2020;29(1):84-93). Several of the networks have made their codes, common data models and analytics software publicly available, such as OHDSI, DARWIN EU®, Sentinel, VAC4EU. This is one of the potential solutions to minimise reproducibility issues in multi-database studies.
Timeliness or speed of running studies is important in order to meet short regulatory timelines, in circumstances where prompt decision-making is needed. Solutions need therefore to be further developed and introduced to be able to run multi-database studies with shorter timelines. Independently from the model used, a critical factor that should be considered in speeding up studies relates to having tasks completed that are independent of any particular study. This includes all activities associated with governance, such as having prespecified agreements on data access, processes for protocol development and study management, and identification and characterisation of a large set of databases. This also includes some activities related to the analysis, such as creating common definitions for frequently used variables, and creating common analytical systems for the most typical and routine analyses. This latter point is made easier with the use of CDMs with standardised analytics and tools that can be re-used to support faster analysis, as demonstrated in DARWIN EU®, where analytical pipelines are being developed to fulfil the needs of EMA-commissioned studies based on pre-specified analysis plans (see Catalogue of Standard Analyses).